Models
1. Logistic Regression Model with / without Regularizations
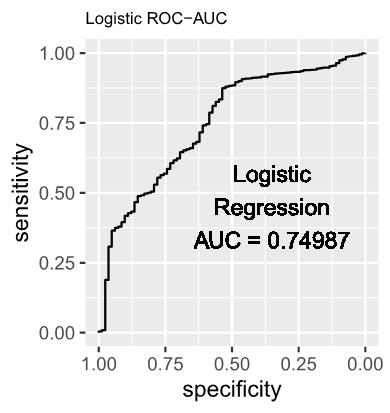
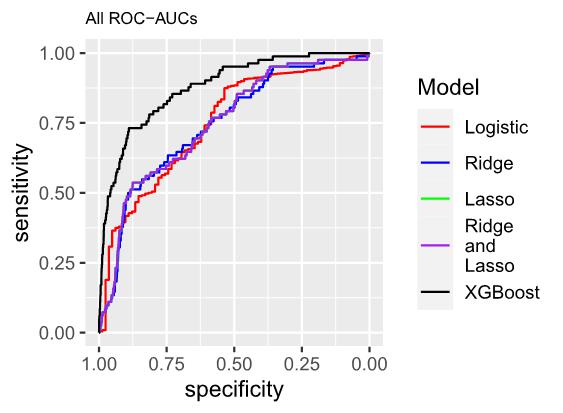
Logistic regression was selected as the baseline model due to its demonstrated effectiveness in accommodating diverse data characteristics. Its appropriateness in the present context lies in its well-established suitability for binary classification tasks, specifically discerning between instances of infected and uninfected status of H5N1 in each county. The mathematical representation of the model is as follows:
\begin{aligned}
Y_i = &\beta_0 +
\beta_1 \cdot \texttt{lat} +
\beta_2 \cdot \texttt{lng} +
\beta_3 \cdot \texttt{month.index} +
\beta_4 \cdot \texttt{avg.temp} +
\beta_5 \cdot \texttt{type(non-poultry)} \\
&+\beta_6 \cdot \texttt{type(poultry)} +
\beta_7 \cdot \texttt{type(wild bird)} +
\beta_8 \cdot \texttt{lat * lng}.
\end{aligned}
The sigmoid transformation below ensures that the predicted probabilities fall within the interval $(0, 1)$, thereby enhancing the model's capability to generate plausible predictions. Serving as a binary classifier, the model is specifically designed to assess the likelihood of H5N1 cases in a county based on the type of outbreak encountered. The application of the sigmoid function contributes to the interpretability and calibration of the model's predictions, aligning them with the probabilistic nature of the logistic regression framework.
\begin{aligned}
&\Pr(Y_i=\texttt{infected} | X_i) = p(X_i) = \frac{e^{\beta_0 + \beta_1 \cdot \texttt{lat} + ... + \beta_8 \cdot \texttt{lat * lng}}}{1 + e^{\beta_0 + \beta_1 \cdot \texttt{lat} + ... + \beta_8 \cdot \texttt{lat * lng}}}\\
&\Pr(Y_i=\texttt{uninfected} | X_i) = 1 - p(X_i) = 1 - \left(\frac{e^{\beta_0 + \beta_1 \cdot \texttt{lat} + ... + \beta_8 \cdot \texttt{lat * lng}}}{1 + e^{\beta_0 + \beta_1 \cdot \texttt{lat} + ... + \beta_8 \cdot \texttt{lat * lng}}}\right).
\end{aligned}
1.1 The estimated log-odds of the Logistic Regression Model is as follows:
\begin{aligned}
\hat{Y}_i = &18.215 -
0.284 \cdot \texttt{lat} +
0.08 \cdot \texttt{lng} -
0.057 \cdot \texttt{month.index} -
0.004 \cdot \texttt{avg.temp} -
0.203 \cdot \texttt{type(non-poultry)} \\
&-0.055 \cdot \texttt{type(poultry)} -
1.997 \cdot \texttt{type(wild bird)} -
0.002 \cdot \texttt{lat * lng}.
\end{aligned}
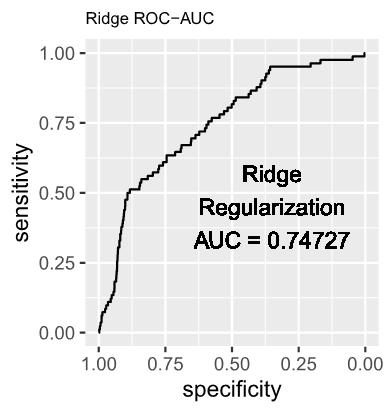
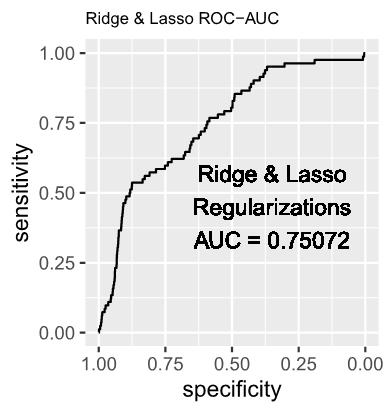
1.2 The estimated log-odds of the Logistic Regression Model with Ridge (L2) Regularization is as follows:
\begin{aligned}
\hat{Y}_i = &9.092 -
0.085 \cdot \texttt{lat} +
0.006 \cdot \texttt{lng} -
0.048 \cdot \texttt{month.index} -
0.001 \cdot \texttt{avg.temp} +
0.043 \cdot \texttt{type(non-poultry)} \\
&+0.168 \cdot \texttt{type(poultry)} -
1.693 \cdot \texttt{type(wild bird)} -
5.818\times 10^{-5} \cdot \texttt{lat * lng}.
\end{aligned}
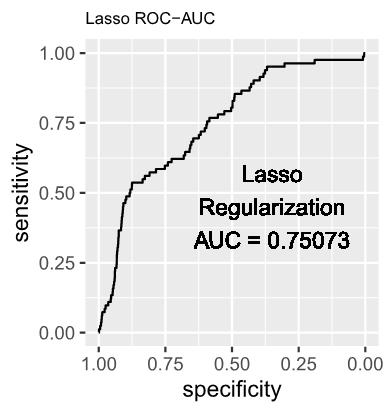
1.3 The estimated log-odds of the Logistic Regression Model with Lasso (L1) Regularization is as follows:
\begin{aligned}
\hat{Y}_i = &17.243 -
0.262 \cdot \texttt{lat} +
0.072 \cdot \texttt{lng} -
0.056 \cdot \texttt{month.index} -
0.004 \cdot \texttt{avg.temp} -
0.197 \cdot \texttt{type(non-poultry)} \\
&-0.049 \cdot \texttt{type(poultry)} -
1.99 \cdot \texttt{type(wild bird)} -
0.002 \cdot \texttt{lat * lng}.
\end{aligned}
1.4 The estimated log-odds of the Logistic Regression Model with Ridge (L2) and Lasso (L1) Regularizations is as follows:
\begin{aligned}
\hat{Y}_i = &17.278 -
0.263 \cdot \texttt{lat} +
0.072 \cdot \texttt{lng} -
0.056 \cdot \texttt{month.index} -
0.004 \cdot \texttt{avg.temp} -
0.197 \cdot \texttt{type(non-poultry)} \\
&-0.05 \cdot \texttt{type(poultry)} -
1.991 \cdot \texttt{type(wild bird)} -
0.002 \cdot \texttt{lat * lng}.
\end{aligned}
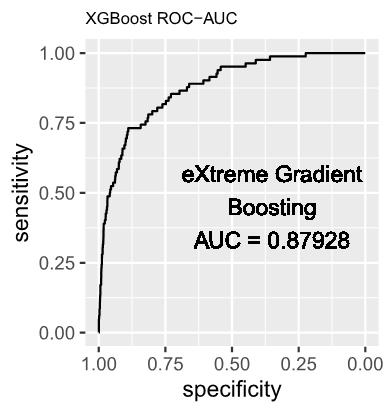
2. eXtreme Gradient Boosting (XGBoost) Model
In contemporary times, XGBoost has emerged as a robust ensemble learning technique, garnering widespread acclaim for its exceptional predictive prowess. The algorithm methodically assembles a set of weak learners, typically manifested as decision trees, and amalgamates their predictions to bolster accuracy and exhibit strong generalization performance on unseen data.
The XGBoost model was trained utilizing the exact tree method, and its performance was systematically monitored through the negative log-likelihood loss at each iteration of the training process. In order to ensure both performance and robustness, a 10-fold cross-validation strategy was implemented at each training round, and certain hyperparameters were systematically adjusted and applied as delineated below:
- The learning rate ($\eta$) was designated as 0.02, serving as the step size shrinkage to mitigate overfitting during the training process.
- The subsample ratio was established at 0.75, indicating that the XGBoost algorithm would stochastically select 75% of the training data for each tree's growth.
- The column subsample ratio for each tree was configured to 0.8, signifying that the XGBoost algorithm would randomly consider 80% of predictors when developing a new tree.
- The maximum depth of the trees was restricted to 10.
- The number of training rounds was predetermined to be 700.
Figure 7: Training and Validation Losses of XGBoost Model over 700 Rounds of Training
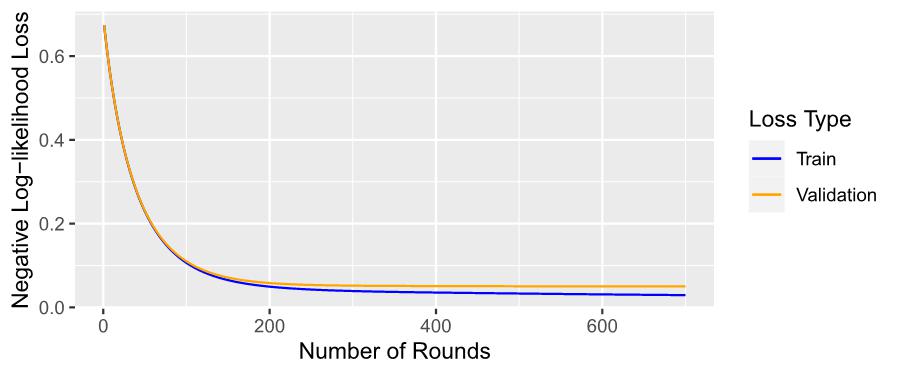
Figure 7 illustrates the progression of training and validation losses across 700 rounds. The training loss, depicted by the blue line, exhibits a consistent and gradual decrease over the course of the training iterations. Similarly, the validation loss, represented by the orange line, also demonstrates a continuous and steady decline.
Figure 8: Feature Importance Plot for XGBoost Model
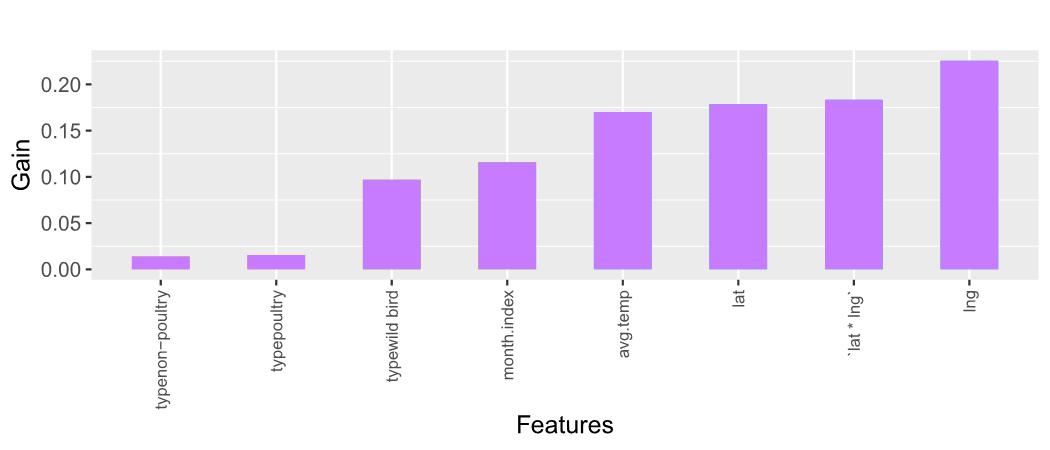
Figure 8 depicts the Gain scores associated with the predictors employed in the XGBoost model. A taller bar, indicative of a higher Gain score, signifies greater importance of the corresponding predictor. Remarkably, pivotal predictors such as lng (longitude), lat * lng (interaction between latitude and longitude), and lat (latitude) have emerged as noteworthy contributors to the predictive efficacy of the model.